CFL Package Quick Start
A handy set of commands that can be copy and pasted to run CFL quickly.
We recommend running cfl from within either a python script (i.e. a .py file) or a Jupyter Notebook. A python script can be easier to run quickly and better enforces reproducibility, but a Jupyter Notebook is more interactive.
1. Import CFL
[1]:
from cfl.experiment import Experiment
2. Load your data as a numpy array
[2]:
# generate random data (replace this with your actual data)
n_samples = 500
import numpy as np
X = np.random.random((n_samples, 20)) # X data has 20 features
Y = np.random.random((n_samples, 3)) # Y data has only 3 features
# print the data shape
# both data sets should be in the shape (n_samples, n_features)
print(X.shape)
print(Y.shape)
(500, 20)
(500, 3)
3. Create parameter dictionaries
[3]:
# Create 3 dictionaries: one for data info, one with CDE parameters, and one with cluster parameters
# the parameters should be passed in dictionary form
data_info = {'X_dims' : X.shape,
'Y_dims' : Y.shape,
'Y_type' : 'continuous' #options: 'categorical' or 'continuous'
}
# pass in empty parameter dictionaries to use the default parameter values (not
# allowed for data_info)
CDE_params = { 'model' : 'CondExpMod',
'model_params' : {
# model architecture
'dense_units' : [50, data_info['Y_dims'][1]],
'activations' : ['relu', 'linear'],
'dropouts' : [0, 0],
# training parameters
'batch_size' : 32,
'n_epochs' : 30,
'optimizer' : 'adam',
'opt_config' : {'lr' : 1e-4},
'loss' : 'mean_squared_error',
'best' : True,
# verbosity
'verbose' : 1,
'show_plot' : True,
}
}
# cluster_params consists of specifying two clustering objects
# CFL automatically recognizes the names of all sklearn.cluster models as keywords
cause_cluster_params = {'model' : 'KMeans',
'model_params' : {'n_clusters' : 4},
'verbose' : 0
}
effect_cluster_params = {'model' : 'KMeans',
'model_params' : {'n_clusters' : 2},
'verbose' : 0
}
4. Set up the CFL pipeline
[4]:
# block_names indicates which CDE and clustering models to use
block_names = ['CondDensityEstimator', 'CauseClusterer', 'EffectClusterer']
# block_params is aligned to block_names
block_params = [CDE_params, cause_cluster_params, effect_cluster_params]
results_path = 'sample_run' # directory to save results to
# Create a new CFL experiment with specified parameters
my_exp = Experiment(X_train=X,
Y_train=Y,
data_info=data_info,
block_names=block_names,
block_params=block_params,
results_path=results_path)
save_path 'sample_run' doesn't exist, creating now.
All results from this run will be saved to sample_run/experiment0000
Block: verbose not specified in input, defaulting to 1
CondExpBase: activity_regularizers not specified in input, defaulting to None
CondExpBase: kernel_regularizers not specified in input, defaulting to None
CondExpBase: bias_regularizers not specified in input, defaulting to None
CondExpBase: kernel_initializers not specified in input, defaulting to None
CondExpBase: bias_initializers not specified in input, defaulting to None
CondExpBase: weights_path not specified in input, defaulting to None
CondExpBase: tb_path not specified in input, defaulting to None
CondExpBase: optuna_callback not specified in input, defaulting to None
CondExpBase: optuna_trial not specified in input, defaulting to None
CondExpBase: early_stopping not specified in input, defaulting to False
CondExpBase: checkpoint_name not specified in input, defaulting to tmp_checkpoints
5. Train the CFL model on the data
[5]:
results = my_exp.train()
#################### Beginning CFL Experiment training. ####################
Beginning CondDensityEstimator training...
No GPU device detected.
Train on 375 samples, validate on 125 samples
Epoch 1/30
375/375 [==============================] - 4s 11ms/sample - loss: 0.7946 - val_loss: 0.7318
Epoch 2/30
375/375 [==============================] - 0s 766us/sample - loss: 0.7251 - val_loss: 0.6690
Epoch 3/30
375/375 [==============================] - 0s 1ms/sample - loss: 0.6614 - val_loss: 0.6112
Epoch 4/30
375/375 [==============================] - 0s 1ms/sample - loss: 0.6032 - val_loss: 0.5587
Epoch 5/30
375/375 [==============================] - 0s 511us/sample - loss: 0.5502 - val_loss: 0.5111
Epoch 6/30
375/375 [==============================] - 1s 2ms/sample - loss: 0.5022 - val_loss: 0.4678
Epoch 7/30
375/375 [==============================] - 1s 1ms/sample - loss: 0.4581 - val_loss: 0.4289
Epoch 8/30
375/375 [==============================] - 0s 1ms/sample - loss: 0.4192 - val_loss: 0.3931
Epoch 9/30
375/375 [==============================] - 0s 922us/sample - loss: 0.3832 - val_loss: 0.3610
Epoch 10/30
375/375 [==============================] - 0s 1ms/sample - loss: 0.3511 - val_loss: 0.3318
Epoch 11/30
375/375 [==============================] - 0s 964us/sample - loss: 0.3223 - val_loss: 0.3055
Epoch 12/30
375/375 [==============================] - 0s 884us/sample - loss: 0.2961 - val_loss: 0.2821
Epoch 13/30
375/375 [==============================] - 0s 899us/sample - loss: 0.2731 - val_loss: 0.2610
Epoch 14/30
375/375 [==============================] - 1s 1ms/sample - loss: 0.2521 - val_loss: 0.2424
Epoch 15/30
375/375 [==============================] - 0s 1ms/sample - loss: 0.2340 - val_loss: 0.2257
Epoch 16/30
375/375 [==============================] - 0s 1ms/sample - loss: 0.2176 - val_loss: 0.2113
Epoch 17/30
375/375 [==============================] - 1s 2ms/sample - loss: 0.2034 - val_loss: 0.1990
Epoch 18/30
375/375 [==============================] - 0s 941us/sample - loss: 0.1917 - val_loss: 0.1878
Epoch 19/30
375/375 [==============================] - 0s 985us/sample - loss: 0.1809 - val_loss: 0.1786
Epoch 20/30
375/375 [==============================] - 0s 1ms/sample - loss: 0.1718 - val_loss: 0.1710
Epoch 21/30
375/375 [==============================] - 0s 1ms/sample - loss: 0.1644 - val_loss: 0.1644
Epoch 22/30
375/375 [==============================] - 0s 874us/sample - loss: 0.1580 - val_loss: 0.1589
Epoch 23/30
375/375 [==============================] - 1s 1ms/sample - loss: 0.1528 - val_loss: 0.1544
Epoch 24/30
375/375 [==============================] - 0s 770us/sample - loss: 0.1483 - val_loss: 0.1508
Epoch 25/30
375/375 [==============================] - 0s 970us/sample - loss: 0.1448 - val_loss: 0.1478
Epoch 26/30
375/375 [==============================] - 0s 709us/sample - loss: 0.1420 - val_loss: 0.1453
Epoch 27/30
375/375 [==============================] - 0s 1ms/sample - loss: 0.1396 - val_loss: 0.1434
Epoch 28/30
375/375 [==============================] - 0s 635us/sample - loss: 0.1377 - val_loss: 0.1417
Epoch 29/30
375/375 [==============================] - 0s 1ms/sample - loss: 0.1361 - val_loss: 0.1405
Epoch 30/30
375/375 [==============================] - 0s 733us/sample - loss: 0.1349 - val_loss: 0.1393
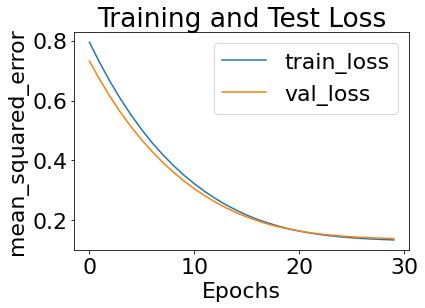
Loading parameters from tmp_checkpoints23032022103422/best_weights
Saving parameters to sample_run/experiment0000/trained_blocks/CondDensityEstimator
CondDensityEstimator training complete.
Beginning CauseClusterer training...
100%|██████████| 500/500 [00:00<00:00, 7144.49it/s]
CauseClusterer training complete.
Beginning EffectClusterer training...
EffectClusterer training complete.
Experiment training complete.
6. Visualizing Results
Refer to the Macrostate Visualization notebook for a more detailed example of how to visualize domain-generic CFL results.
[8]:
# visualize cause side macrostates
from cfl.visualization.basic_visualizations import visualize_macrostates
# visualize cause macrostates
visualize_macrostates( data=X,
feature_names=None,
cause_or_effect='cause',
exp_path=my_exp.get_save_path(),
data_series='dataset_train',
subtract_global_mean=True)
# visualize effect macrostates
visualize_macrostates( data=Y,
feature_names=None,
cause_or_effect='effect',
exp_path=my_exp.get_save_path(),
data_series='dataset_train',
subtract_global_mean=True)
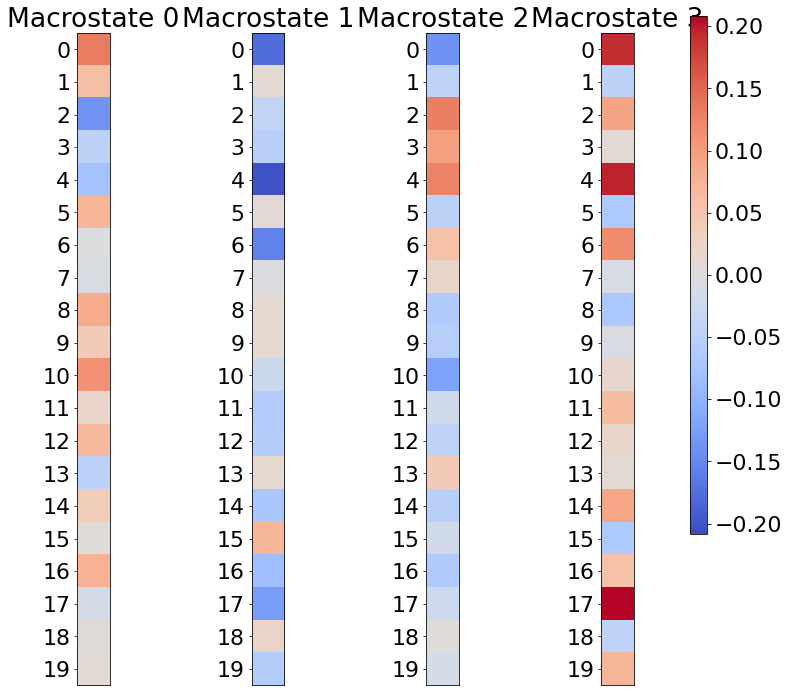
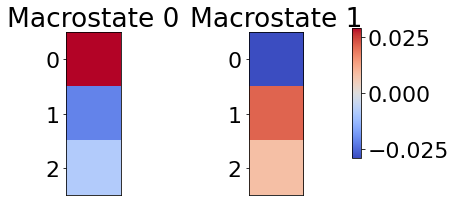
7. Tuning CFL
Once you’ve set up a CFL pipeline, the hyperparameters needs to be tuned as is the case with any other machine learning model.
To tune the CondDensityEstimator, you can tune the hyperparamters specified in CDE_params by following the Optuna tutorial or using your technique of choice. You can also consider different methods of conditional density estimation altogether (available models can be found in cfl.cond_density_estimation, and instructions for adding your own models can be found in the Adding Blocks tutorial).
To tune the CauseClusterer and/or EffectCLusterer, hyperparameters can be tuned using CFL’s built-in method described in the clusterer tuning tutorial, or with your technique of choice. You can also consider different clustering algorithms altogether, (all Sklearn clustering models are visible to CFL, and instructions for adding your own models can be found in the Adding Blocks tutorial).